Integrated Antibody Sequence and Structure
Management, Analysis and Prediction
A fully integrated antibody discovery system
Version 3.4.1
Companies are welcome to use this public version of abYsis, but need to be aware that this is not a secure server. After trialing the system, companies wishing to install a local version of abYsis, which can also store and analyse proprietary sequence and 3D structure data, may request a commercial licence.
For further information please contact our specialist distributor: Chemogenomix. You may also see further information on the UCL Business e-licensing website.
This public version of abYsis is made available largely through the generous support of commercial licensees.
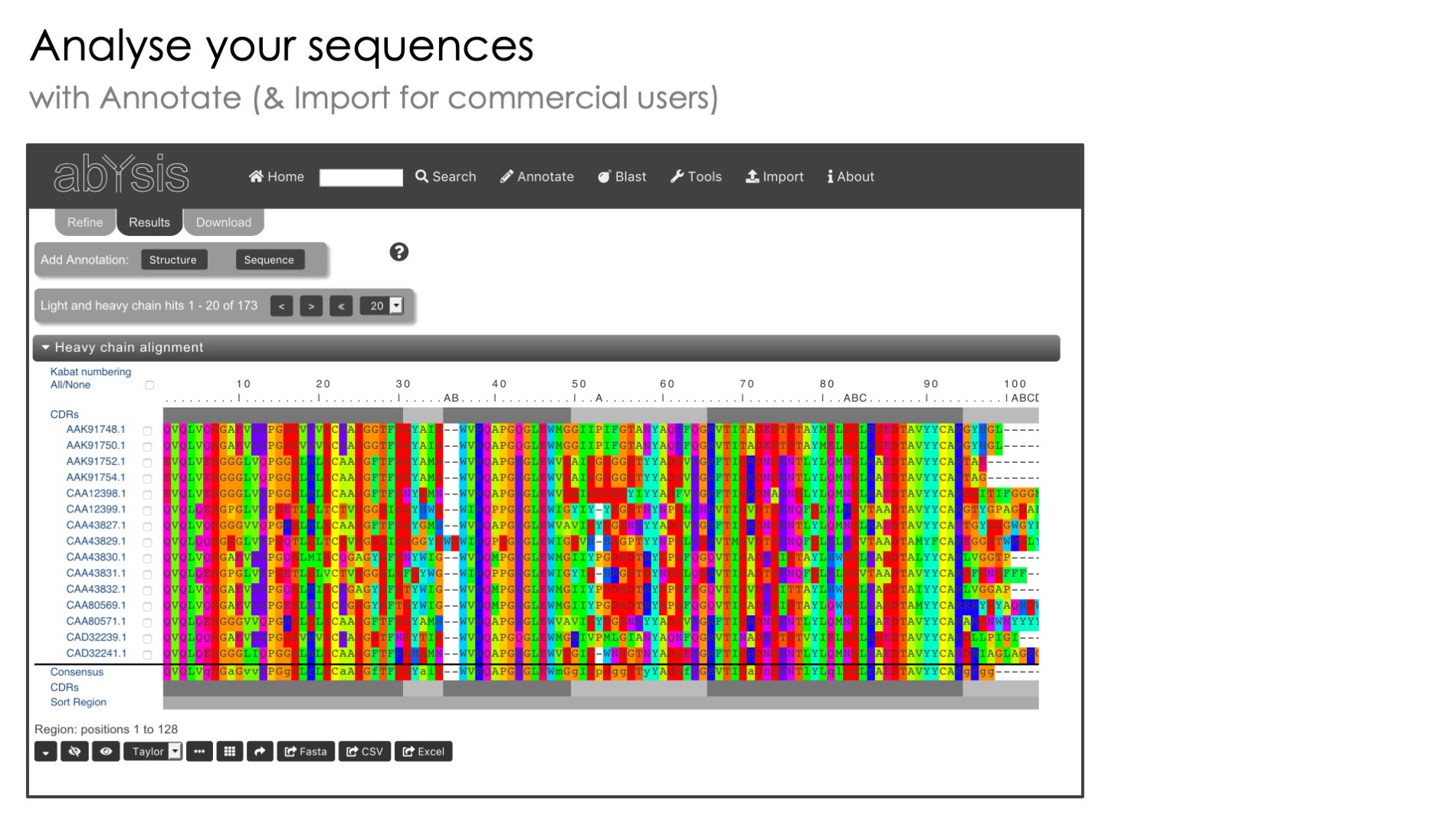
Antibody Sequence Management
Pipeline reads data files in standard formats such as EMBL, PDB and FASTA
3D structure integration
Intelligent processing of 3D data from Protein Data Bank
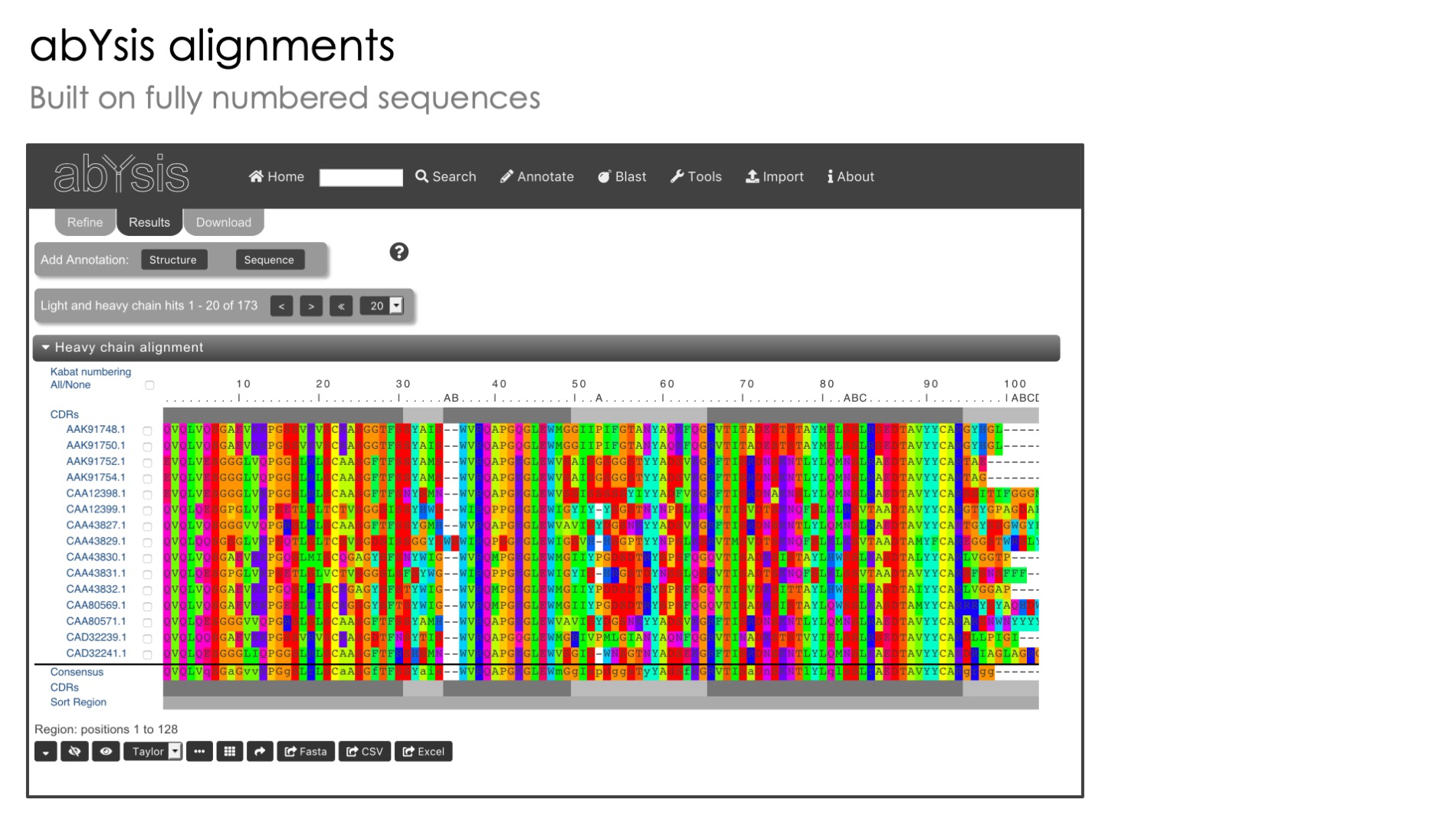
Automated Antibody Numbering
Standardised numbering of antibody sequences based on Kabat and other popular methods
Relational Database Storage
PostgreSQL database storage of all imported data for flexible analysis
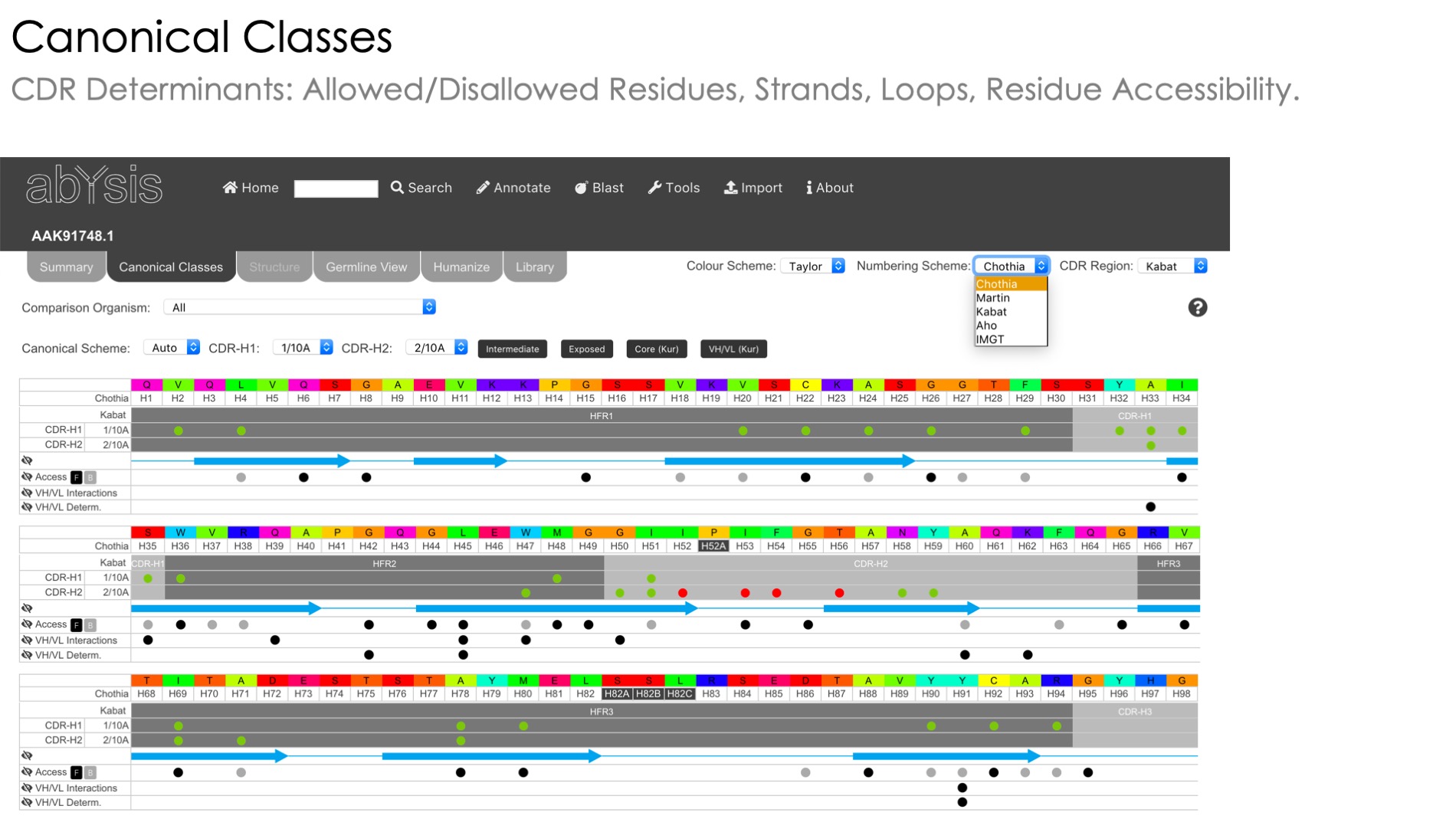
Canonical Class Annotation
Prediction of canonical class based on published methods
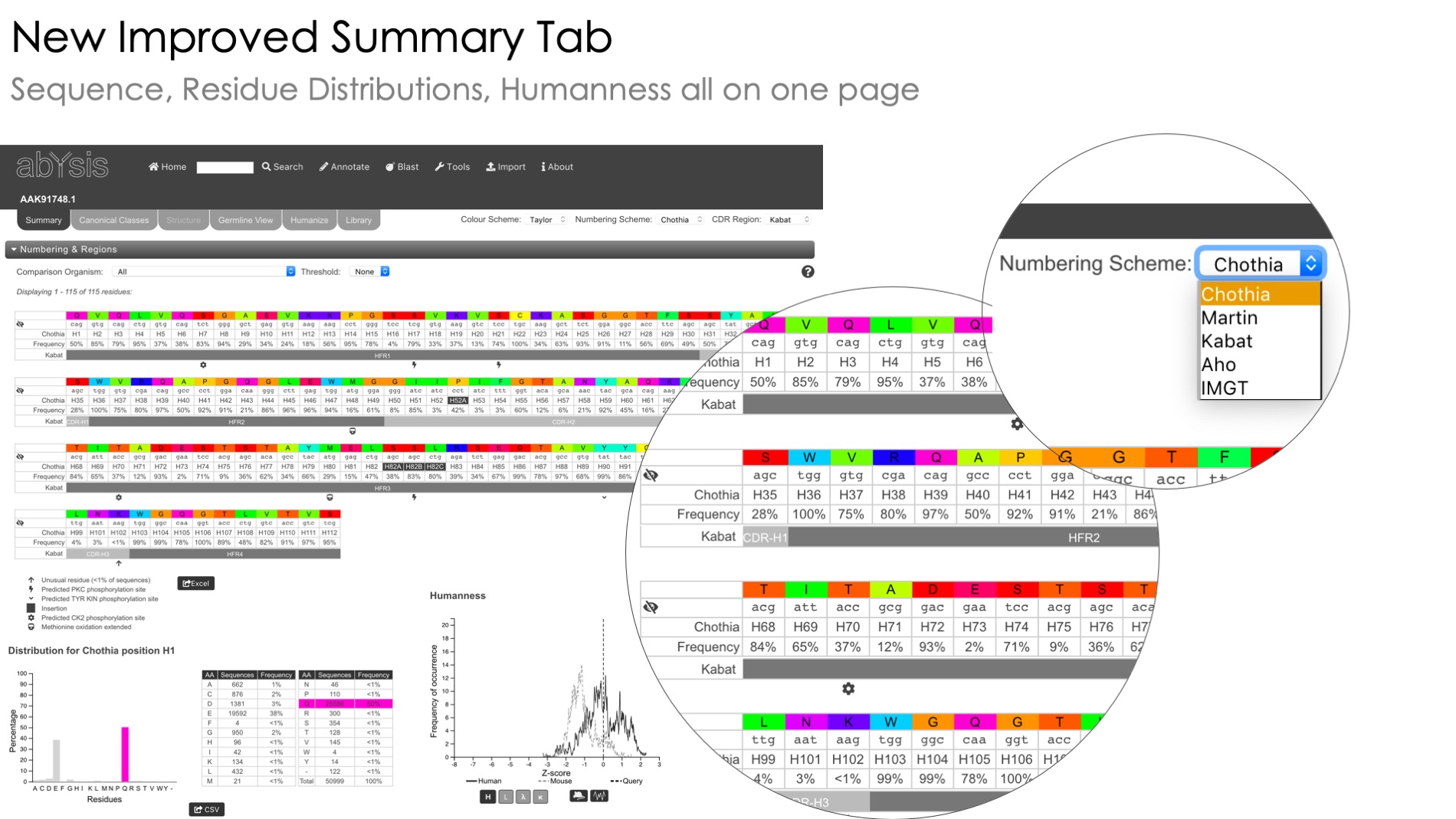
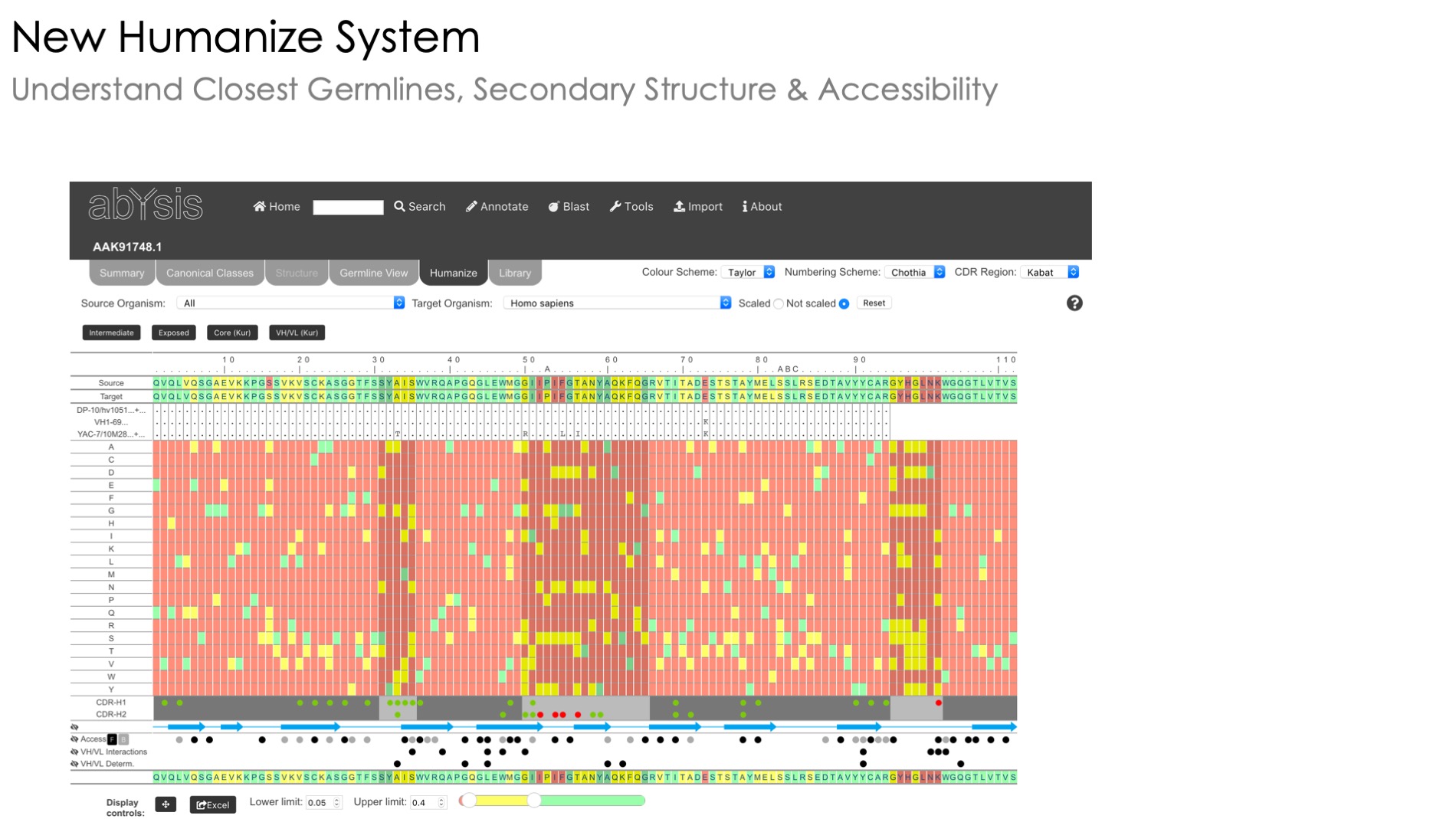
Humanness assessment
Estimate humanness of sequences under investigation
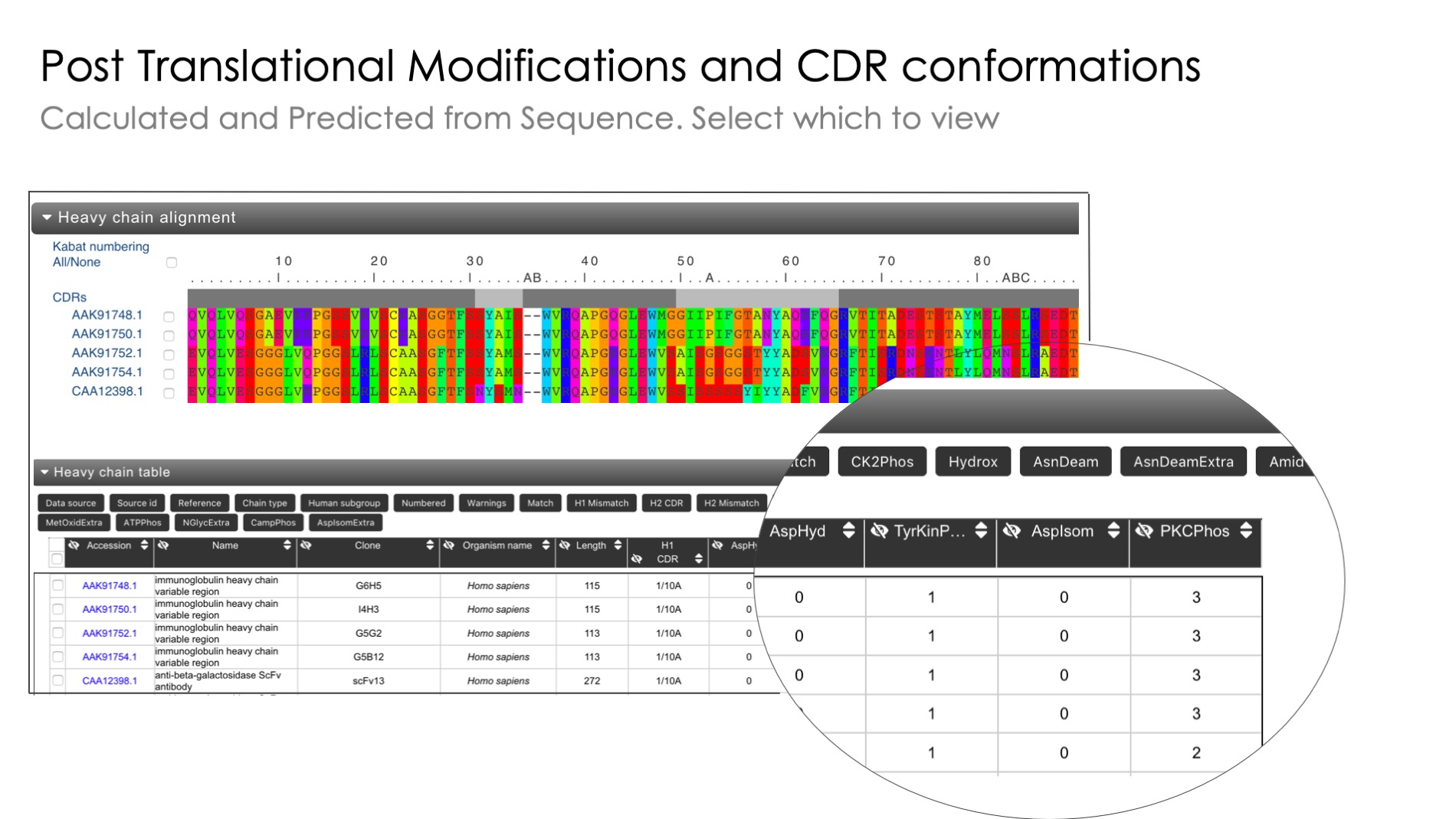
Unusual residue identification
Identification of unusual residues
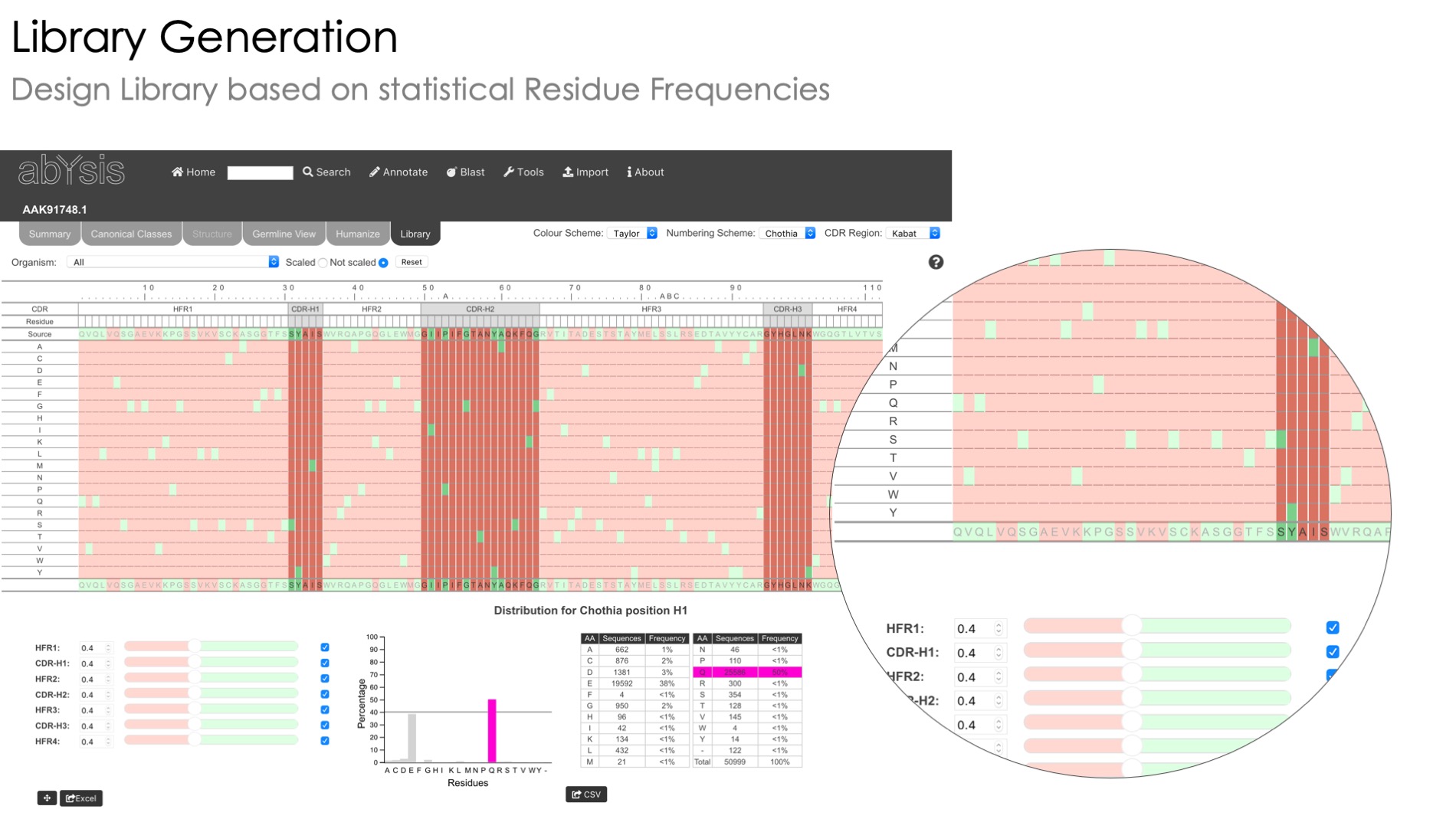
Humanization
A heatmap view of unusual residues to help you humanize your sequence
DNA/protein Alignment
Link DNA sequence to protein sequence with gene modeling tools
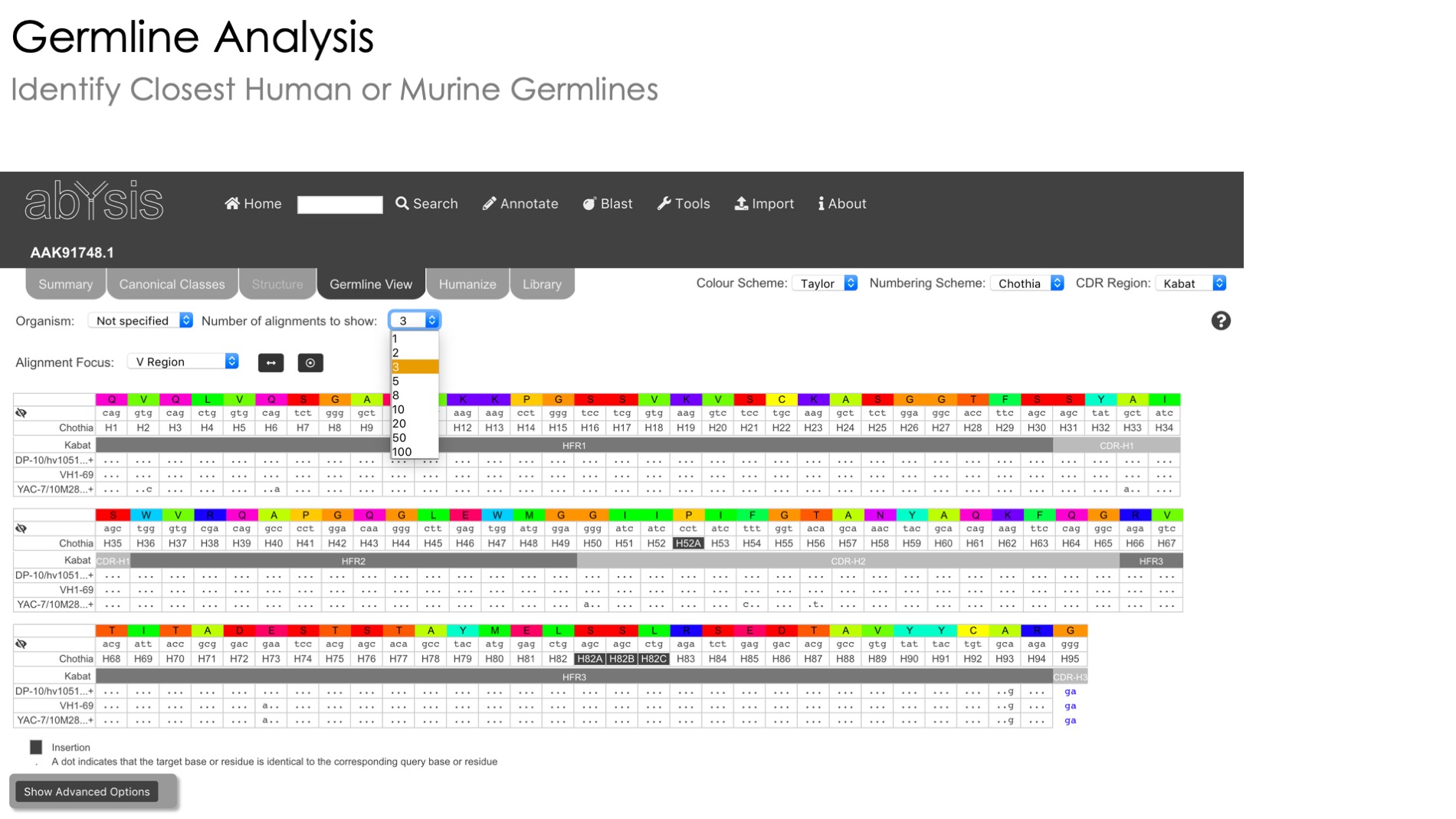
Germline View
Investigate the genomic underpinnings of your sequence
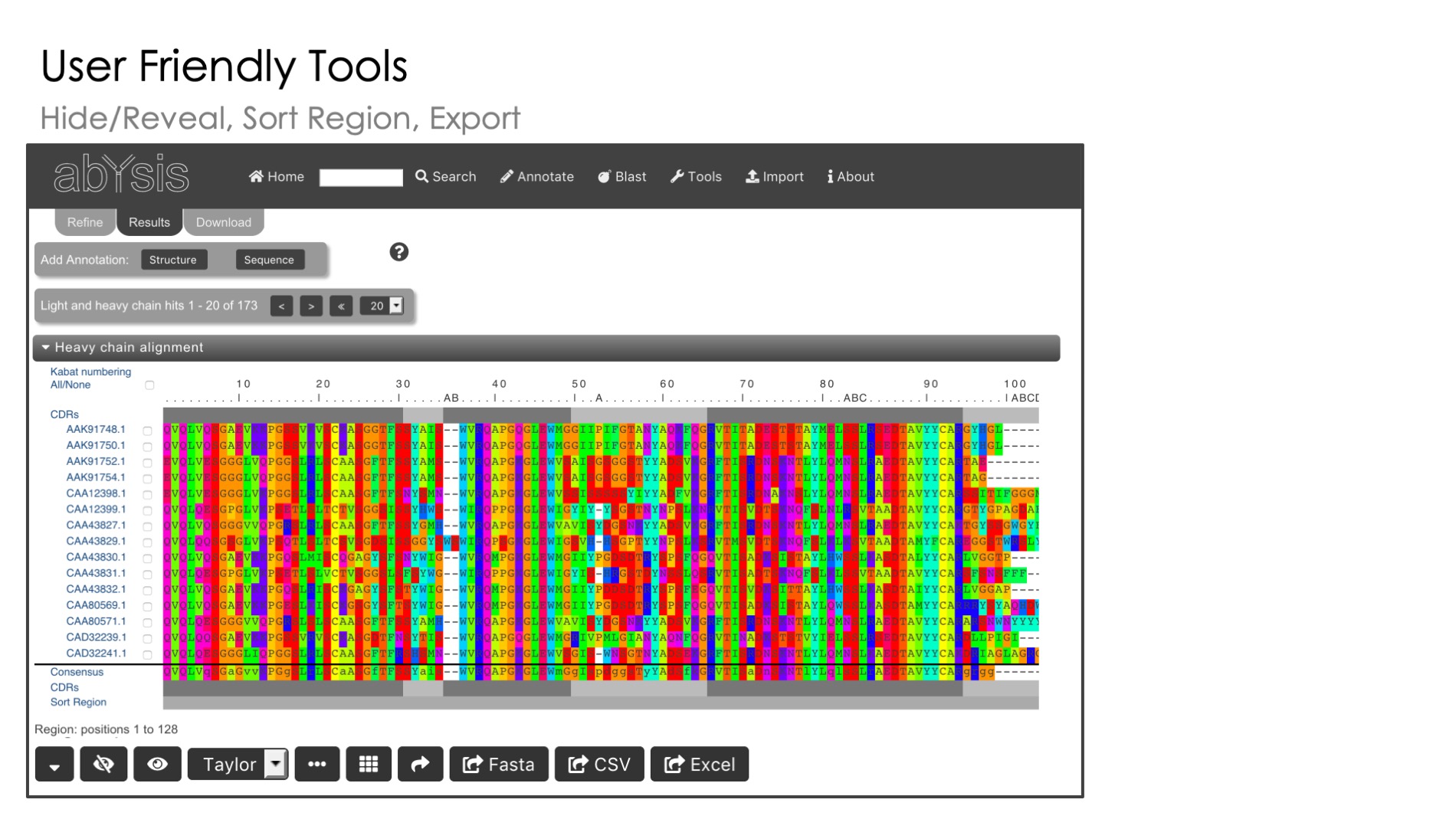
User Input
Use our flexible sequence input tools to calculate all main features on the fly
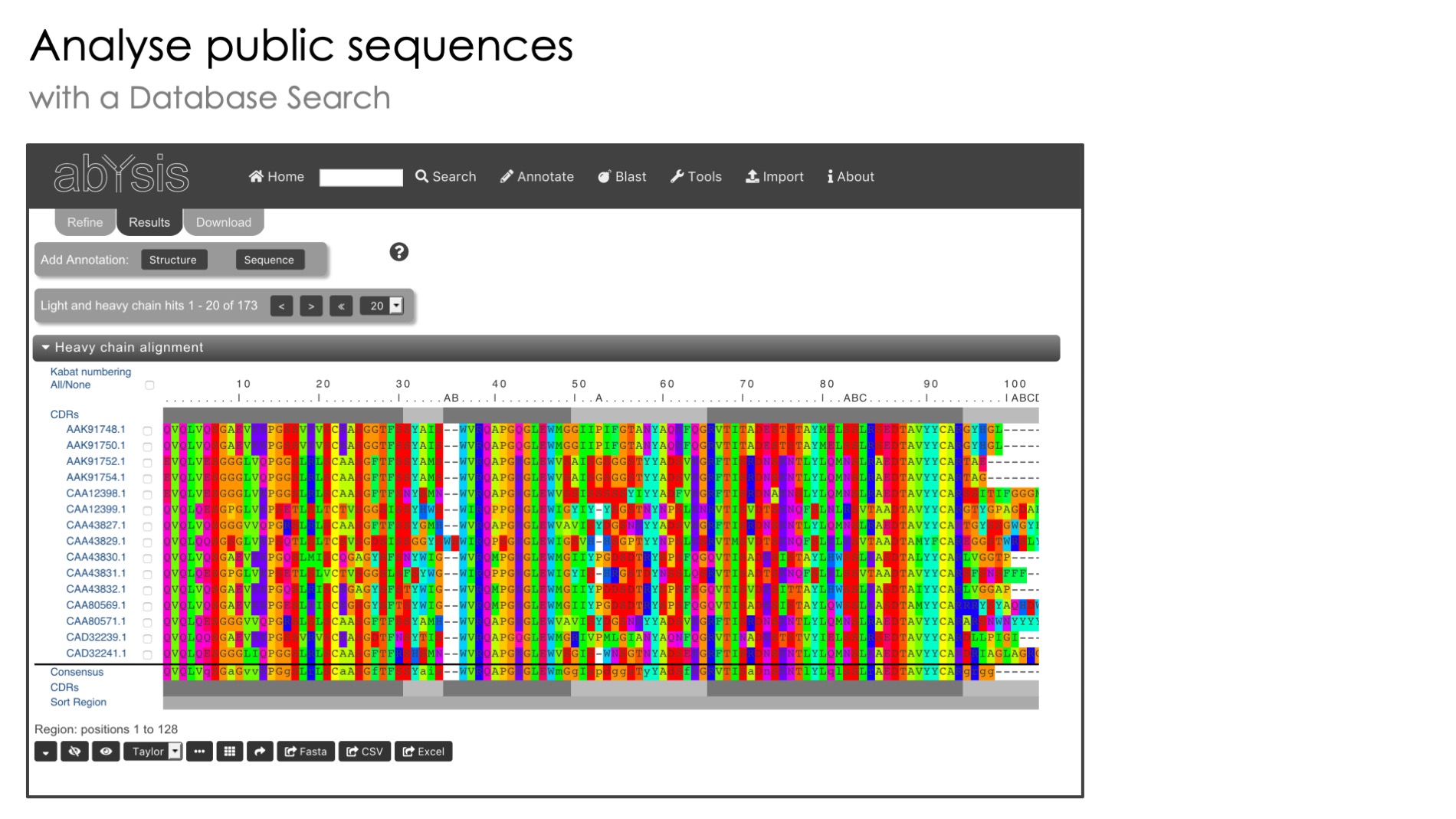
Perform a quick homology search of abYsis antibody sequences using the Blast algorithm.
Query sequences can be either DNA or Protein. Database is only protein sequences.
The Blast server is located either on your own internal installation of abYsis (if licensing a commercial version) or on the public abYsis server (if using the free public version).
Hit sequences are all available within abYsis for deeper analysis.
Do you have sequences that are not in abYsis?
Select the Annotate option from the top menu and paste in your sequences (either DNA or protein).
abYsis will automatically number each sequence and then generate all desired properties, such as unusual residues, humanness and germline, as well as accessing the Humanize tool.
My sequence will not number in the annotate tool. Why is that?
Here are the most frequent issues:
Your sequence is too short so we are unable to number a fraction of an antibody. (Frequently with DNA from sequencing experiments).
Your sequence is unusual and the algorithm cannot find any of the initial motifs it expects in order to seed the numbering (Are you using a less common organism?)
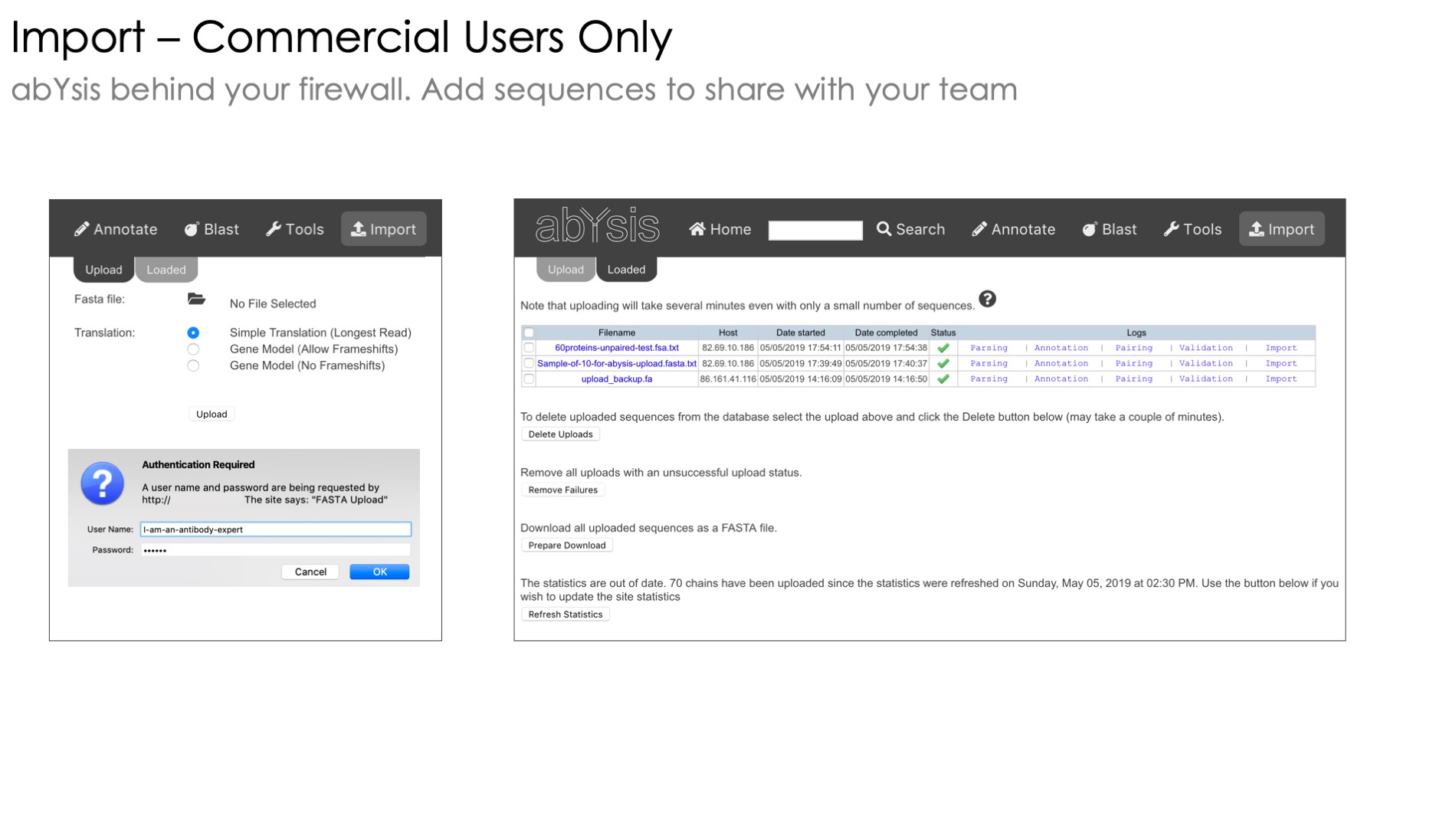
Only available with the commercial version of abYsis, this part of the system lets you annotate batches of sequences and store them for later use without having to re-enter them*
Combine the accession, organism and data source fields to identify different sets of sequences within your project as well as to quickly distinguish your projects from your colleagues.
Here are the 6 key items to put in the header of your fast file, each separated by the | character
Accession,
Partner Accession (for paired heavy and light chains)
DNA or Protein sequence
Species,
Data Source,
Chain_Type Heavy/Light
*When your abYsis system is updated to a new version all sequences must be reprocessed including your own.